Difference between revisions of "Bioreactor"
Darren Roos (Talk | contribs) |
Darren Roos (Talk | contribs) m |
||
| (5 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | [[Image: | + | [[Image:bioreactor_picture1.jpg|center|400px]] |
==System description== | ==System description== | ||
| Line 18: | Line 18: | ||
==Current documentation== | ==Current documentation== | ||
| − | [Here] is the current lab manual/documentation containing the: wiring information, process diagram, operating procedure and software installation information | + | [https://drive.google.com/open?id=1xzLvDu-u5TOpixQoFqj68_FGfRugsTrS Here] is the current lab manual/documentation containing the: wiring information, process diagram, operating procedure and software installation information |
==Current software setup== | ==Current software setup== | ||
| Line 63: | Line 63: | ||
A useful article for understanding how it works can be found [https://towardsdatascience.com/the-unscented-kalman-filter-anything-ekf-can-do-i-can-do-it-better-ce7c773cf88d here]. | A useful article for understanding how it works can be found [https://towardsdatascience.com/the-unscented-kalman-filter-anything-ekf-can-do-i-can-do-it-better-ce7c773cf88d here]. | ||
| − | The report for the project can be found [[ | + | The report for the project can be found [[Media:darren_roos_bioreactor_report.pdf|here]]. |
Code for the project can be found [https://github.com/darren-roos/CML_code here]. | Code for the project can be found [https://github.com/darren-roos/CML_code here]. | ||
| − | The documentation compiled by Sphinx can be found [[ | + | The documentation compiled by Sphinx can be found [[Media:darren_roos_cml_code_docs.pdf|here]]. |
| − | The lab manual and documentation can be found [here] | + | The lab manual and documentation can be found [https://drive.google.com/open?id=1xzLvDu-u5TOpixQoFqj68_FGfRugsTrS here] |
| − | + | ||
==Future work== | ==Future work== | ||
Latest revision as of 16:07, 24 November 2019
Contents
System description
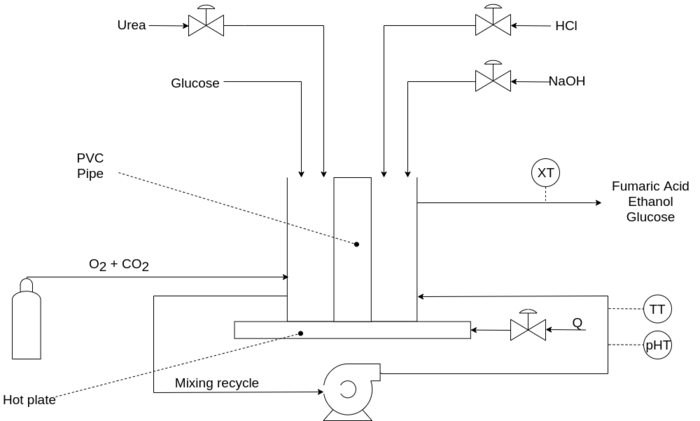
Fumaric acid and ethanol are produced through the aerobic fermentation of glucose by Rhizopus oryzae. Ethanol is an unwanted by-product and its concentration is reduced by sparging with carbon dioxide. There are two phases in the process: a growth phase and production phase. During the growth phase, the fungus is grown on a PVC pipe within the cylindrical fermentation vessel with high concentrations of urea in a batch process. Thereafter, in the production phase, the vessel is rinsed out and continuous fermentation begins with lower concentrations of urea.
Mixing in the vessel is caused by the continuous recycling of the reactor fluid from the base to the head of the reactor and as such the system is modelled as a CSTR. The direction of the recycle switches periodically to prevent the build-up of fungus and subsequent clogging of the pump. Volume is kept constant through the use of an overflow. The overflow's fumaric acid, glucose and ethanol concentrations is measured.
Current documentation
Here is the current lab manual/documentation containing the: wiring information, process diagram, operating procedure and software installation information
Current software setup
The current system has Labview and Python code. Labview is used to interface with the hardware, display readings, and control the temperature. There are currently two closed loops in the system, one for the temperature and one for the pH. The temperature loop consists of a discrete PID controller and pulse width modulator that manipulates temperature by turning the heating plate element on and off. The pH controller is part of the pH probe and its workings are proprietary.
The Python code currently runs on Python (v3.7) with the following packages:
- numpy (v1.16.4)
- pandas (v0.24.2)
- scipy (v1.3.0)
- filterpy (v1.4.5)
- matplotlib (v3.1.0)
- tqdm (v4.32.1)
it contains code that makes it easy for a different parts of the system to be replaced. There is code that can simulate a model that interacts with either offline data from the bioreactor or online data from the reactor. The code contains it's own plotting code and does not interface with Labview's plotting code. An Unscented Kalman Filter is used for state estimation in the system.
Previous work
Andre Naude (2017)
On the bio side much work has been done by Naude who did his PhD on the reactor in 2018. The three published papers relating to his work can be found here, here, and here. His full PhD thesis can be found here
Reuben Swart (2019)
Swart also worked on the bio side and has completed his Masters on the reactor. His work can be found [here]
Darren Roos (2019)
Roos worked on the control side. He characterized the pH probe that is used in the reactor in terms of its accuracy, linearity, drift and dynamic response. The probe (if recently calibrated) is both accurate to within 0.045% and linear throughout the range of 2 pH to 11 pH. 24h drift experiments found that the average drift of the measurement is 0.01375 pH per hour. Dynamic tests found a first order system with a gain of one and time constant of 130 seconds described the response well.
He also developed a nonlinear model of the system that is used in state estimation. An Unscented Kalman Filter is used in the state estimation system. A useful article for understanding how it works can be found here.
The report for the project can be found here. Code for the project can be found here. The documentation compiled by Sphinx can be found here. The lab manual and documentation can be found here
Future work
- Translation of the Labview code into Python would make future development easier because it is impractical to do state estimation and advanced control in Labview. A potentially good resource for interfacing with the hardware can be found here
- An improved model of the system that includes actual metabolic pathway and regime information. This would likely have to build on work done on the bio side to determine the exact mechanisms involved.
- Add controllers for the fumaric acid and ethanol concentration. This would involve a good understanding of how to manipulate these variables.